Revolutionizing Gene Editing: Retron Library Recombineering Explained
Written on
Chapter 1: Introduction to Gene Editing Innovations
The CRISPR-Cas9 gene-editing system has transformed the landscape of medical science. This conventional method operates by identifying and cutting out defective genes responsible for diseases, essentially replacing them with healthy gene sequences. However, this "cut-and-paste" approach is often criticized for being invasive, with potential for unintended consequences.
To address these concerns, researchers have been exploring a gentler alternative. A team from the Wyss Institute for Biologically Inspired Engineering at Harvard University and Harvard Medical School has developed a groundbreaking technique known as Retron Library Recombineering (RLR). This innovative method simplifies the gene-editing process while minimizing invasiveness.
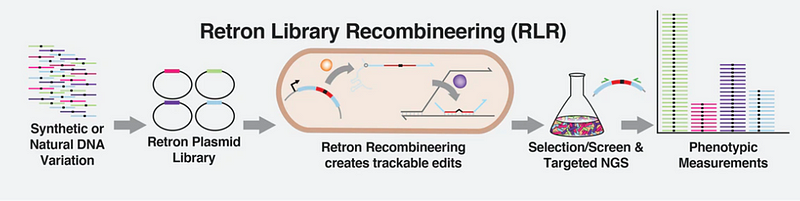
Section 1.1: What is Retron Library Recombineering?
RLR can generate millions of mutations at once and tags these mutant cells with "barcodes," allowing for simultaneous screening of the entire pool. This capability facilitates the generation and analysis of vast amounts of data. Researchers advocate that RLR is a more adaptable gene-editing tool, reducing the toxicity linked with CRISPR. Retrons, which are segments of bacterial DNA, undergo reverse transcription to create fragments of single-stranded DNA (ssDNA).

“This new synthetic biology tool brings genome engineering to an even higher level of throughput, which will undoubtedly lead to new, exciting, and unexpected innovations.”
~ Don Ingber, Lead Researcher
Subsection 1.1.1: The Mechanism Behind Retrons
Originally recognized by scientists in the 1980s, it wasn't until last year that researchers discovered that ssDNA produced by retrons can detect viral infections in cells, acting as a component of the bacterial immune response. This characteristic, once considered a peculiar feature of bacteria, now offers precise and flexible gene editing capabilities in bacteria, yeast, and even human cells.

Integrating ssDNA with the desired mutation into an organism's genome can occur in two primary ways. The first involves using CRISPR to insert the mutant sequence during the DNA repair process. The second, and more desirable method, incorporates a single-stranded annealing protein (SSAP) into a replicating cell, allowing the SSAP to integrate the mutant strand into the DNA of daughter cells.
Section 1.2: Advantages of Using RLR
The unique advantage of retrons lies in their ability to produce ssDNA within cells without damaging the native DNA. This contrasts with traditional methods that often force DNA into cells externally. Another benefit is the tracking capability of retron sequences, functioning like barcodes to monitor which cells were edited and the effects of those edits.
Following some initial setbacks and genetic adjustments to the bacteria, the research team successfully edited populations of E. coli. They discovered that over 90% of the population had integrated the desired sequence after just 20 generations. The barcoding nature of retrons allowed for straightforward tracking of which edits successfully transferred the desired genes into the bacterial genome, greatly expediting the process.
Chapter 2: Applications of RLR in Genetic Research
The researchers took their investigation further by applying RLR to randomly fragmented DNA sourced from an E. coli strain resistant to an antibiotic. These fragments were utilized to create a library of tens of millions of genetic sequences contained within retron sequences in plasmids.
Consequently, this novel tool holds the potential to develop new beneficial strains and address challenges like antibiotic resistance, all while being a safer alternative to CRISPR.
The video titled "Retron Library Recombineering vs. CRISPR" delves deeper into the intricacies of these gene-editing techniques and their implications for the future of genetic engineering.

Complete findings were published in the Journal of PNAS.
Stay informed with the content that matters — Join my mailing list.